SARS-CoV-2 lineage portraits
Lineage Portraits is a custom visualization tool I created during the COVID-19 pandemic for illustrating and comparing the mutations behind viral lineages in circulation. The tool was built for and used on a weekly basis by leading experts in biology, virology, epidemiology and public health at The Massachusetts Consortium on Pathogen Readiness (MassCPR) led by Harvard Medical School.
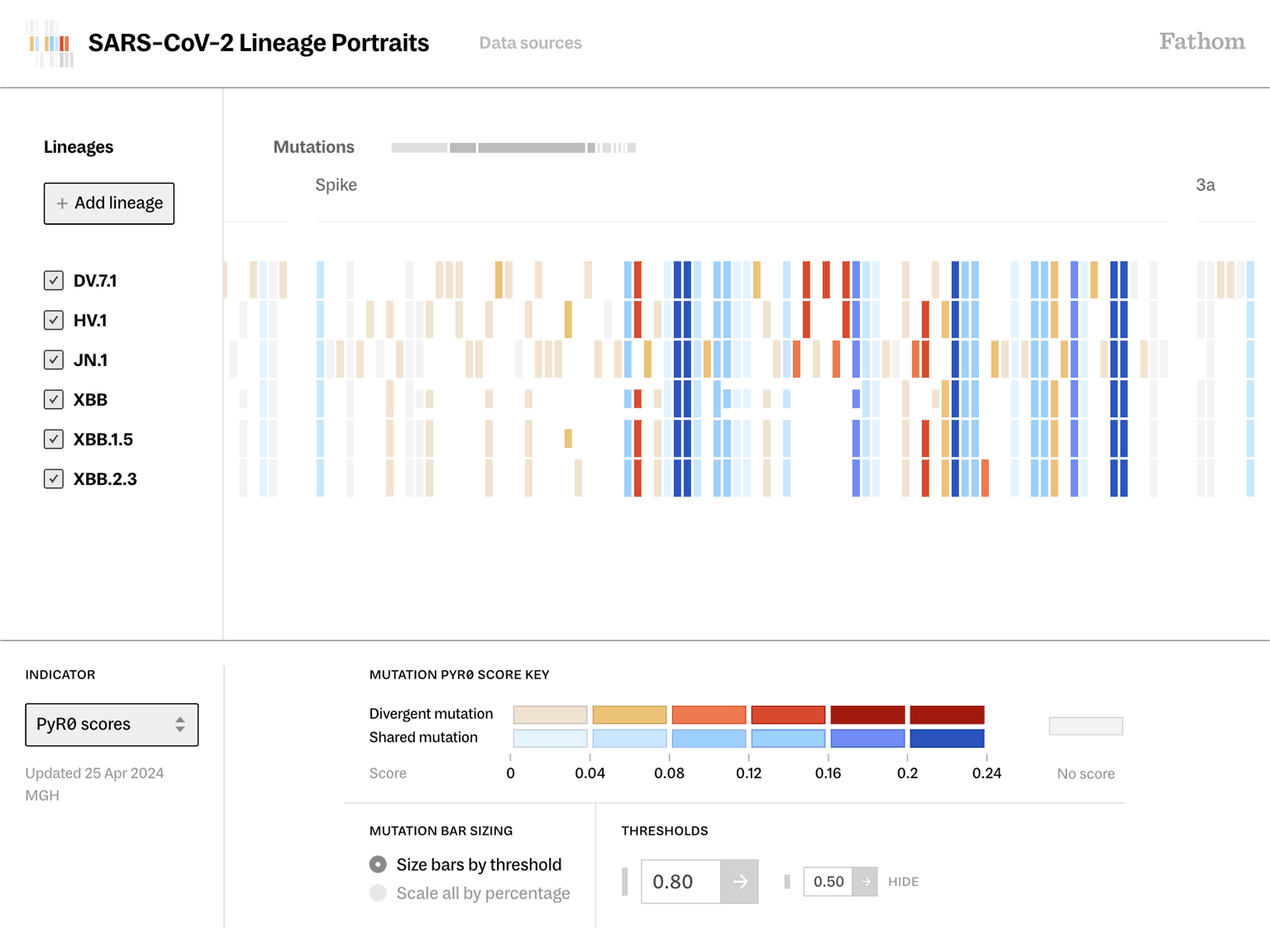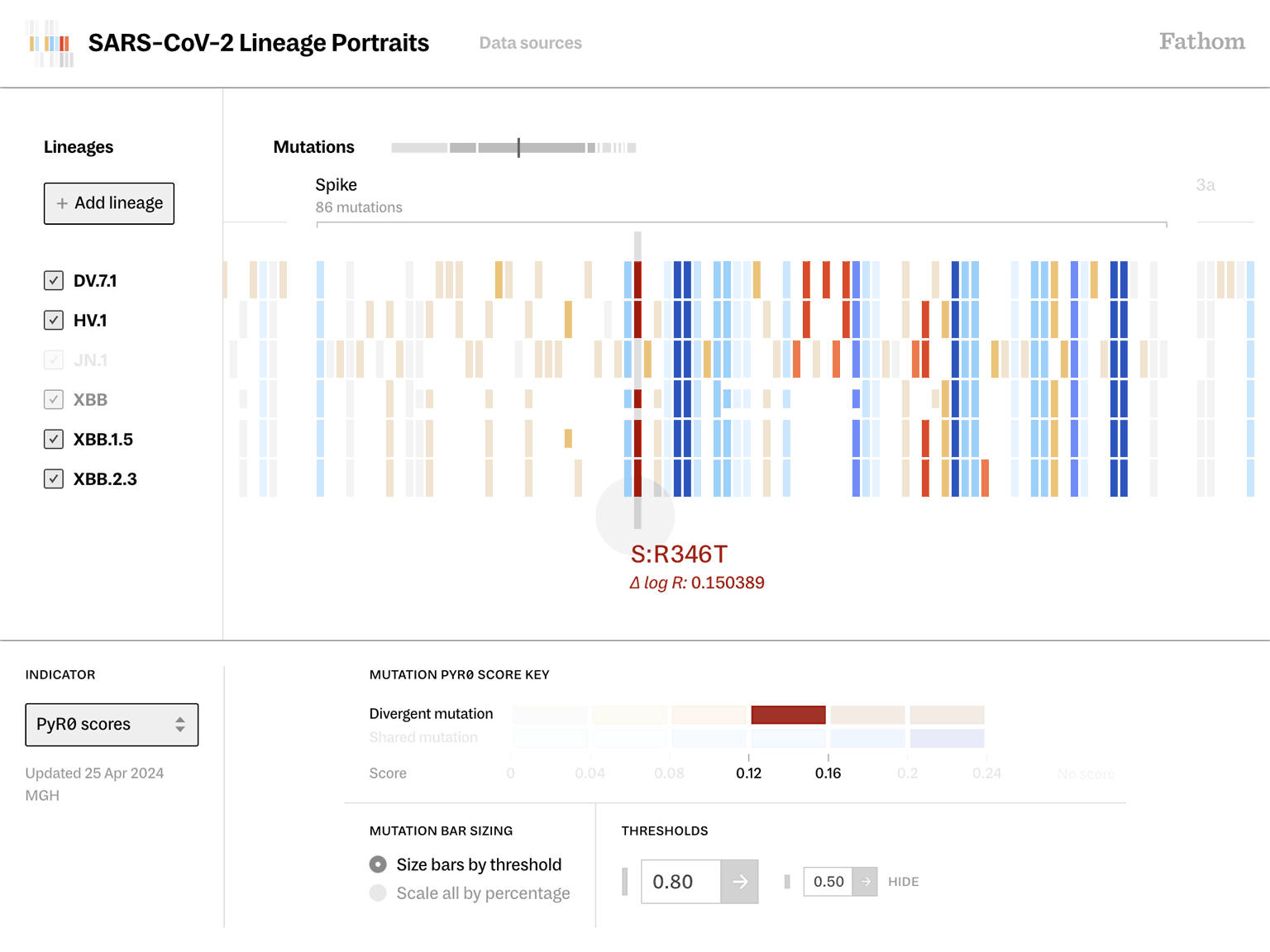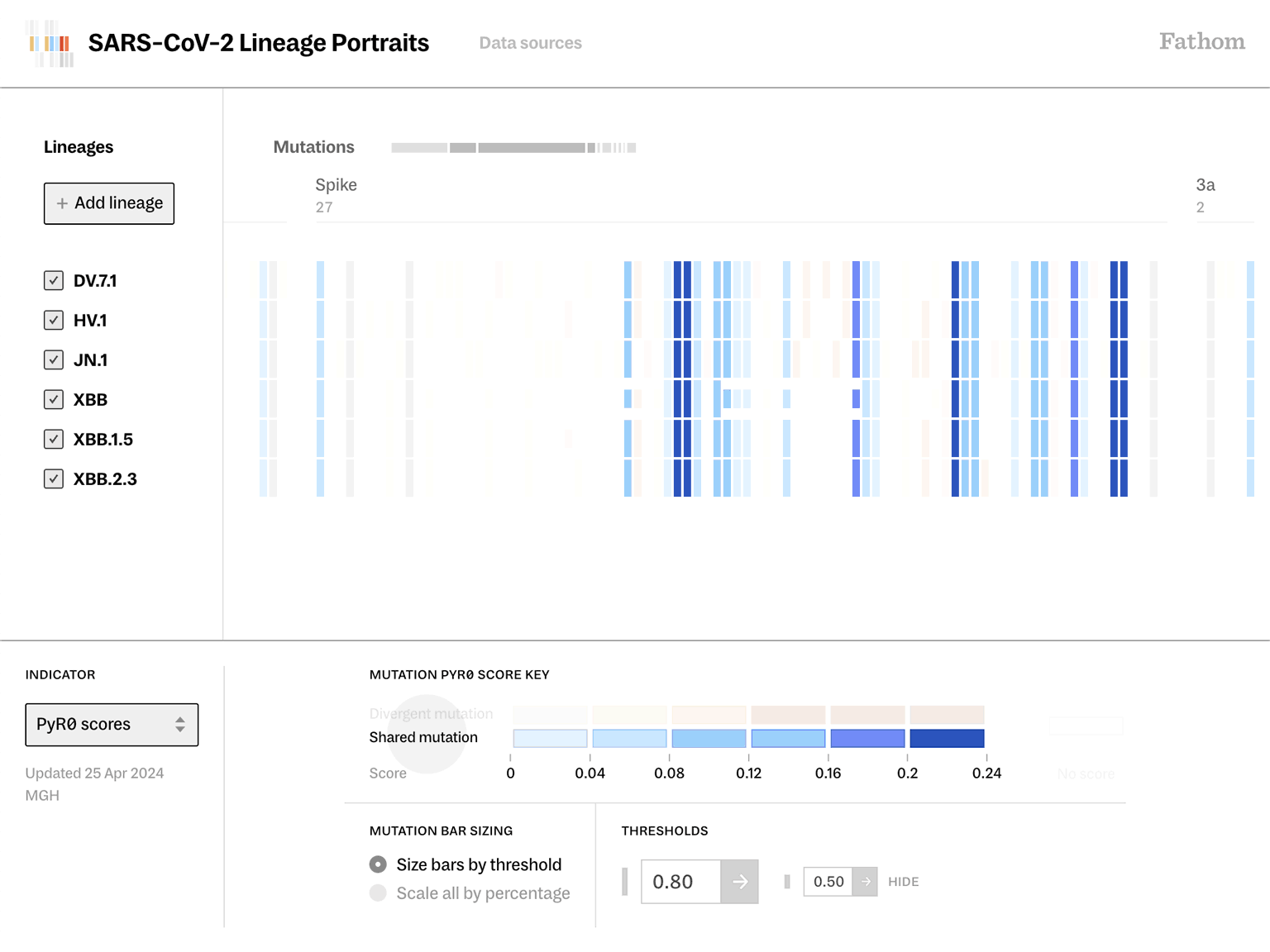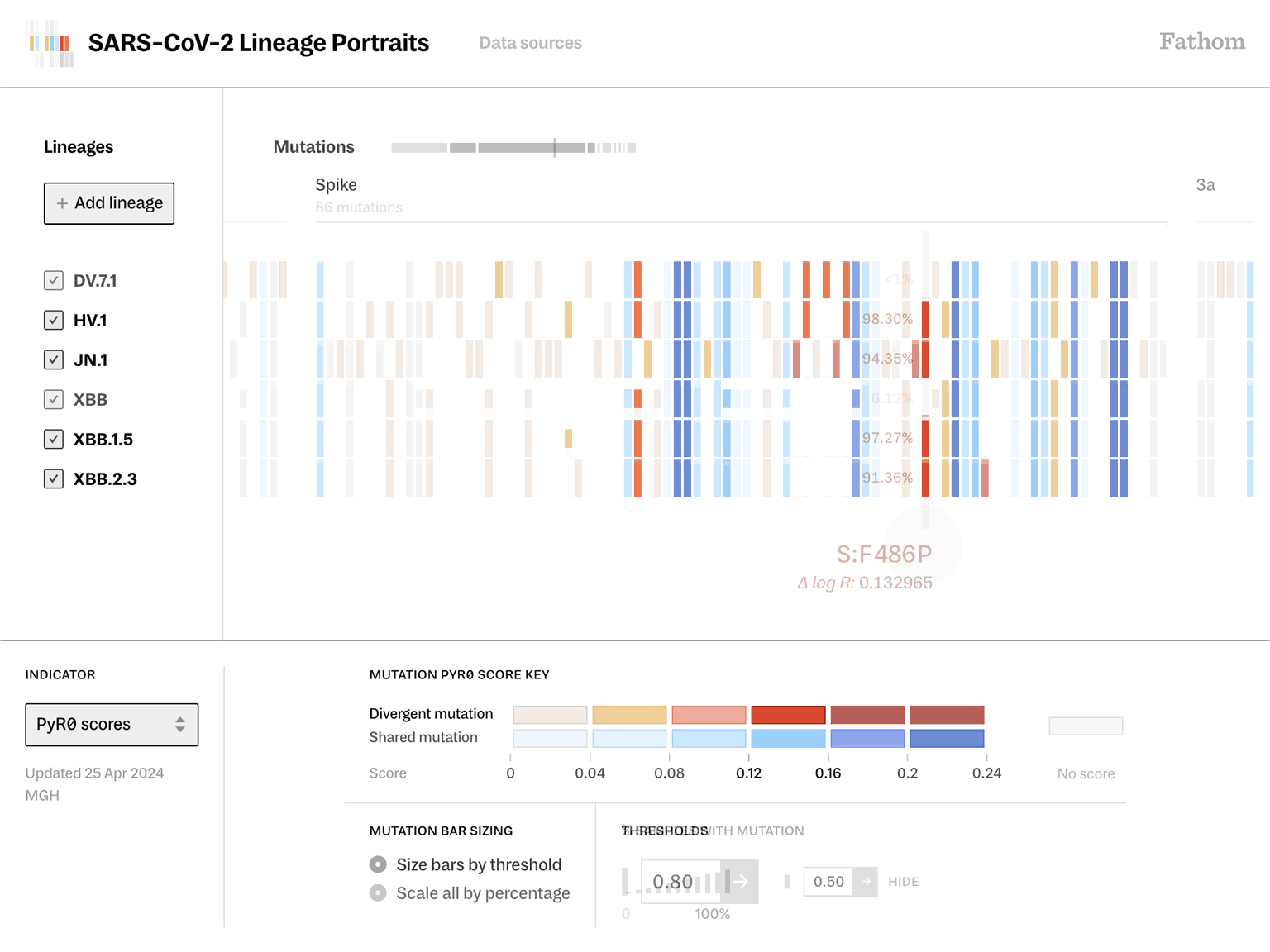
As a globally active virus like SARS-CoV-2 evolves, the classification systems to identify and label new viral lineages have become even more complicated than the problem they’re intended to solve. Additionally, standard visual representations of genomic data lack the information hierarchy needed to assess the relative importance of how clusters of mutations are evolving.
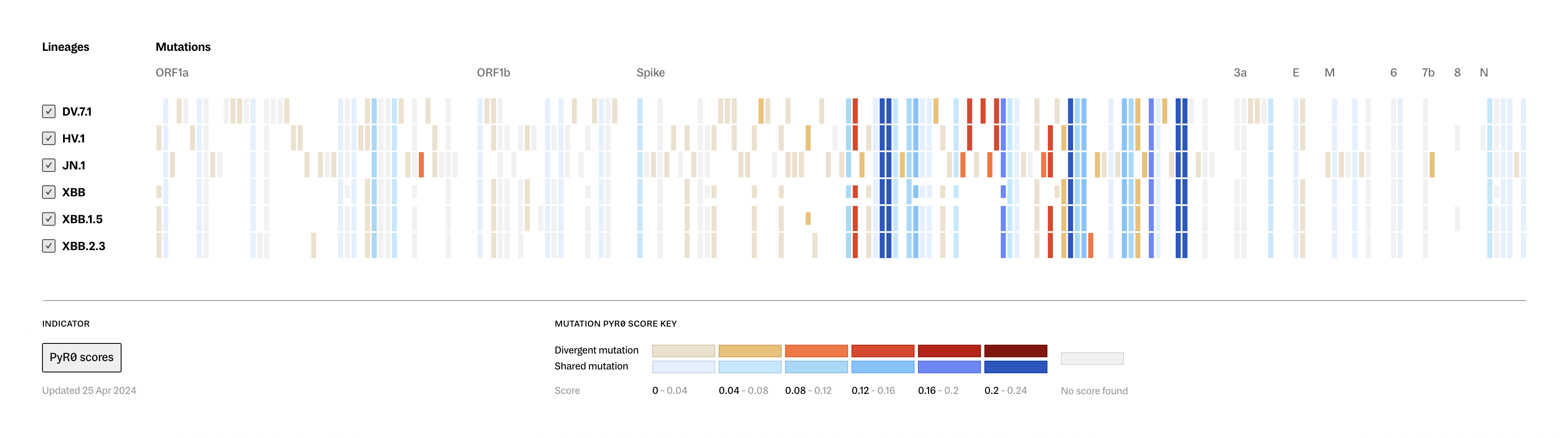
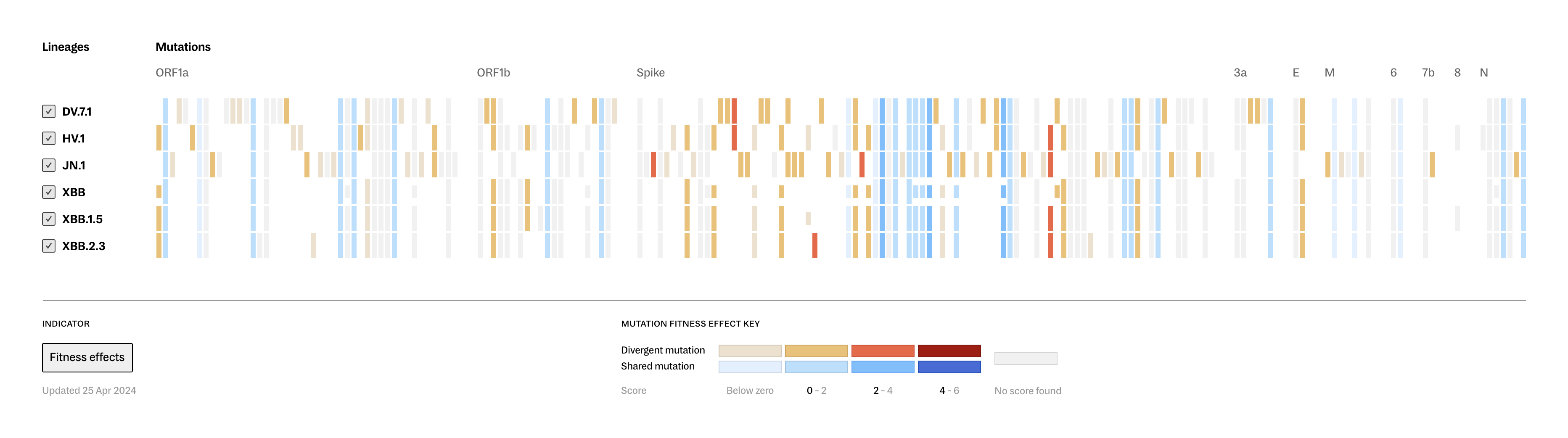
Lineage Portraits is a more useful reference point for those research efforts. It provides an ‘at a glance’ representation of any lineage for direct comparison. The mutations present in each lineage are visualized as a row of colored blocks, with statistical scoring methods applied to highlight more critical sets of mutations with warmer colors.
Showing these patterns in the aggregate provides researchers a way to rapidly compare which aspects of new lineages diverge from established ones. The data is updated as new samples are sequenced, and the tool serves as a resource that can be further annotated as a screenshot and added to a slide deck for sharing.
Lineage Portraits is part of Banyan, a suite of software for outbreak preventation that we’ve developed at Fathom as part of the Sentinel project in coordination with the Sabeti Lab at the Broad Institute of Harvard and MIT.
Font in use: MD System